Recently, the Vegetable Molecular Design Breeding Team at the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, published a research paper, entitled “Regional Active Transcription Associates with Homoeologous Exchange Breakpoints in Synthetic Brassica Tetraploids” in the journal Plant Physiology. The study depicted multi-omics profiles of the early generations of newly synthesized allopolyploids, revealing the active transcriptional characteristics at homoeologous exchange (HE) breakpoints.

Polyploidization plays a crucial role in plant evolution and trait development, contributing significantly to species differentiation, diversity formation, and crop domestication. Synthetic polyploids are one of the effective means to promote crop diversity and are increasingly applied in crop breeding. Previous research has shown that chromosomal rearrangements and aneuploidy frequently occur in synthetic polyploids, accompanied by the remodeling of epigenetic patterns. However, the mechanisms underlying this phenomenon and their impact on gene expression and plant phenotypes remain unknown.
This study utilized a research system involving allopolyploids obtained by distant hybridization between Brassica rapa and Brassica oleracea, followed by genome doubling. Through multi-omics analysis of the leaves of the parents, F1 hybrids, and polyploid offspring over several years, the study depicted the dynamic changes in genomic fragments, gene transcription, and methylation modifications during the early generations of newly synthesized polyploids (Figure 1).
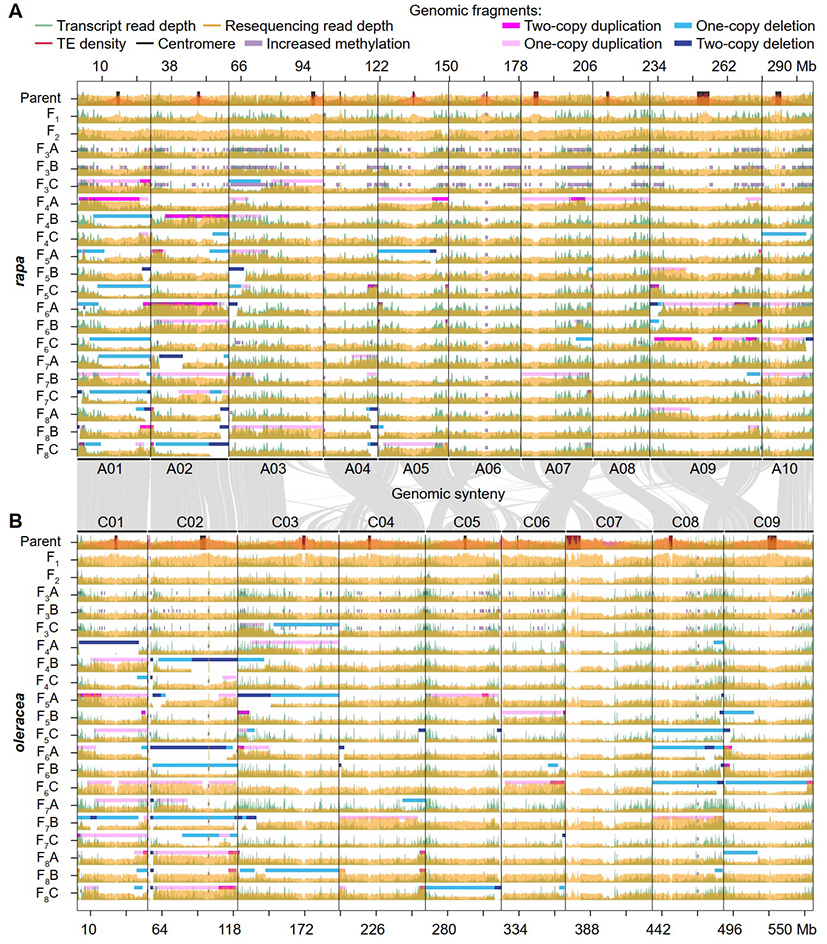
Fig. 1 The landscape of genome structure, gene expression and DNA methylation variations.
The research found that large HE events frequently occurred in newly synthesized allopolyploids after self-crossing, with their frequency positively correlated with the level of collinearity between the two subgenomes. These HE events, resulting from polyploid self-crossing, not only disrupted the genetic consistency and stability of the polyploid offspring but also created materials with genetic diversity. Additionally, the findings suggest that in research utilizing HE to introduce exogenous fragments, high-frequency introgression can be achieved by consecutive self-crossing for 1-2 generations after distant hybridization. Genes located within HE fragments exhibited significant dosage effects in their expression levels, but no significant changes in methylation levels were detected within these HE fragments. Although the overall genome methylation levels fluctuated across different generations, these changes did not significantly impact gene expression. Further research revealed that HE breakpoint positions mostly occurred within genes. These genes, along with neighboring genes, did not show significant functional bias but indicated active transcriptional characteristics in terms of gene sequence length, surrounding TE content, transcription, and methylation levels (Figure 2).
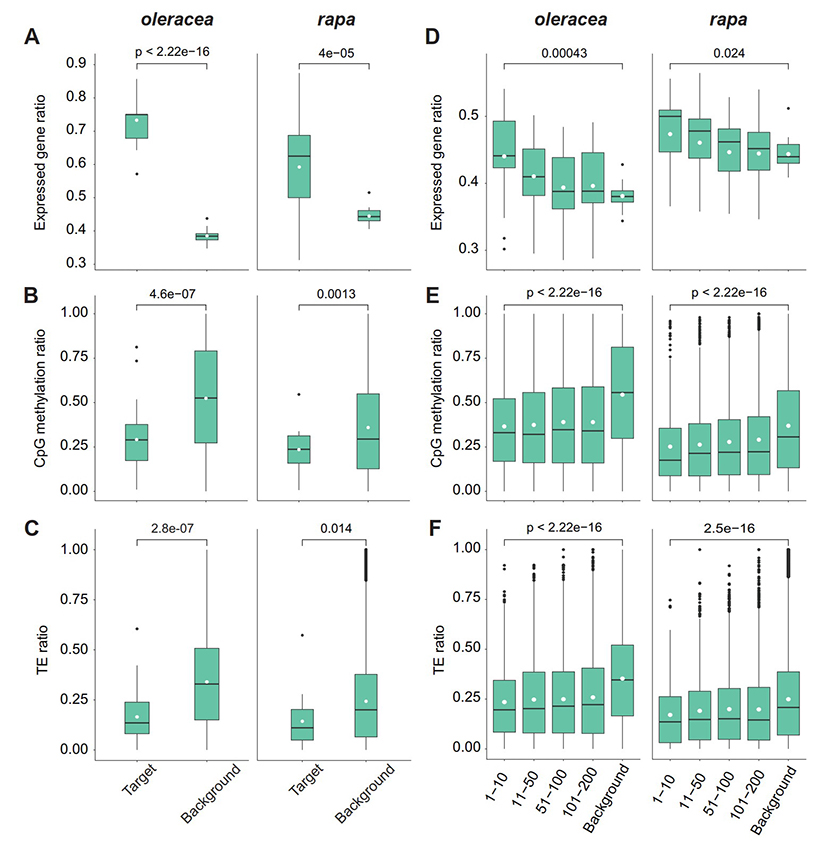
Fig 2. The characteristics of HE breakpoints.
Assistant Researcher Chengcheng Cai, PhD graduate Yinqing Yang (currently at Shandong Agricultural University), and Associate Researcher Lei Zhang from the Vegetable Molecular Design Breeding Innovation Team at the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, are co-first authors of this paper. Professor Feng Cheng, Professor Xiaowu Wang, Professor Kang Zhang, and Professor Michael Freeling from the University of California, Berkeley, are co-corresponding authors. Several teachers and graduate students from the Vegetable Molecular Design Breeding Team participated in this work. This work was supported by the National Natural Science Foundation of China, Central Public-interest Scientific Institution Basal Research Fund, Innovation Program of the Chinese Academy of Agricultural Sciences, and Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture and Rural Affairs, P.R. China.
The link for the paper: https://doi.org/10.1093/plphys/kiae434