Mutations within cis-regulatory regions have less pleiotropic effects than those in protein-coding areas, and thus are ideal targets to fine-tune gene expression and create customized phenotypes. Recently, numerous efforts to identify and characterize cis-regulatory elements (CREs) in plants have been made. However, those identified CREs only underly a small fraction of phenotypic variations and environmental responses, and thus limiting the applications of CREs for crop improvement. To discover and characterize the vast unexplored regulatory sequences at a large scale is still necessary to comprehensively understand and utilize CREs.

The botanical family Cucurbitaceae (cucurbits) includes a number of economically important species, particularly those bearing edible fruits such as cucumber, melon and watermelon. Their inferior ovary can develop into fruit, sharing a similar biological process. Some homologous genes showing conserved functions have been characterized for fruit development of cucurbit crops. Taken together, we expect that some conserved regulatory elements should also be involved in the conserved biological process of fruit development. The availability of increasing genome assemblies of cucurbits provides an unprecedented opportunity to exploit CREs at the genome level.
Recently, the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences and Qingdao Agricultural University published a research paper titled "Identification and functional characterization of conserved cis-regulatory elements responsible for early fruit development in cucurbit crops" in The Plant Cell. In this study, 392,438 conserved non-coding sequences (CNSs) were identified, including 82,756 that are specific to cucurbits, by comparative genomics. Genome-wide profiling of accessible chromatin regions (ACRs) and gene expression patterns mapped 20,865–43,204 ACRs and their potential target genes for two fruit tissues at two key developmental stages in six cucurbits. Integrated analysis of CNSs and ACRs revealed 4,431 syntenic orthologous CNSs, including 1,687 cucurbit-specific CNSs that overlap with ACRs that are present in all six cucurbit crops and that may regulate the expression of 757 adjacent orthologous genes. CRISPR mutations targeting two CNSs present in the 1,687 cucurbit-specific sequences resulted in substantially altered fruit shape and gene expression patterns of adjacent NAC1 (NAM, ATAF1/2 and CUC2) and EXT-like (EXTENSIN-like) genes, validating the regulatory roles of these CNSs in fruit development. These results not only provide a number of target CREs for cucurbit crop improvement, but also provide insight into the roles of CREs in plant biology and during evolution.
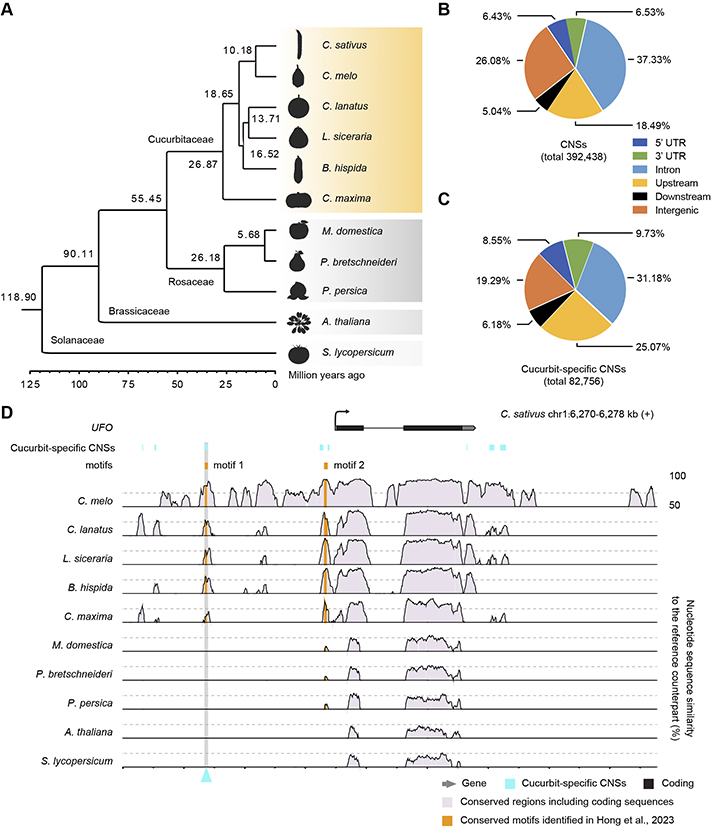
Identification of conserved non-coding sequences (CNSs)
Doctor Hongjia Xin from the Institute of Vegetables and Flowers of Chinese Academy of Agricultural Sciences is the first author, and Professor Zhang Zhonghua from Qingdao Agricultural University is the corresponding author of the paper. This work was supported by the State Key Laboratory of Vegetable Biobreeding, National Natural Science Foundation of China, China National Key Research and Development Program for Crop Breeding, National High-level Talent Special Support Program in China, the “Taishan Scholar” Foundation of the People’s Government of Shandong Province.
Paper link: https://doi.org/10.1093/plcell/koae064