Spinach is a dioecious plant, making it an ideal vegetable crop for studying plant sex determination and sex chromosome evolution. The study of spinach genome sequence and genetic variation can facilitate the identification and utilisation of genes for important agronomic traits in spinach. Compared with shorter variant types (SNP, Indel), longer structural variants (SV) can capture more genetic information and regulate agronomically important traits more significantly. However, fewer structural variants have been studied in spinach, which inhibits the rapid mining of genes related to important agronomic traits in spinach.

Recently, Qian Wei's team from the Spinach Genetic Breeding Group of the Institute of Vegetable and Flower Research, Chinese Academy of Agricultural Sciences (CAAS) published a research paper entitled "Pan-genome analysis of 13 Spinacia accessions reveals structural variations associated with sex chromosome evolution and domestication traits in spinach" in the Plant Biotechnology Journal. This study constructed the spinach pan-genome for the first time, uncovered important structural variations and revealed their roles in sex chromosome evolution and domestication traits in spinach. This will lay the foundation for further research on spinach sex chromosome evolution, mining important agronomic trait-related genes, and targeted breeding of high-quality spinach varieties.
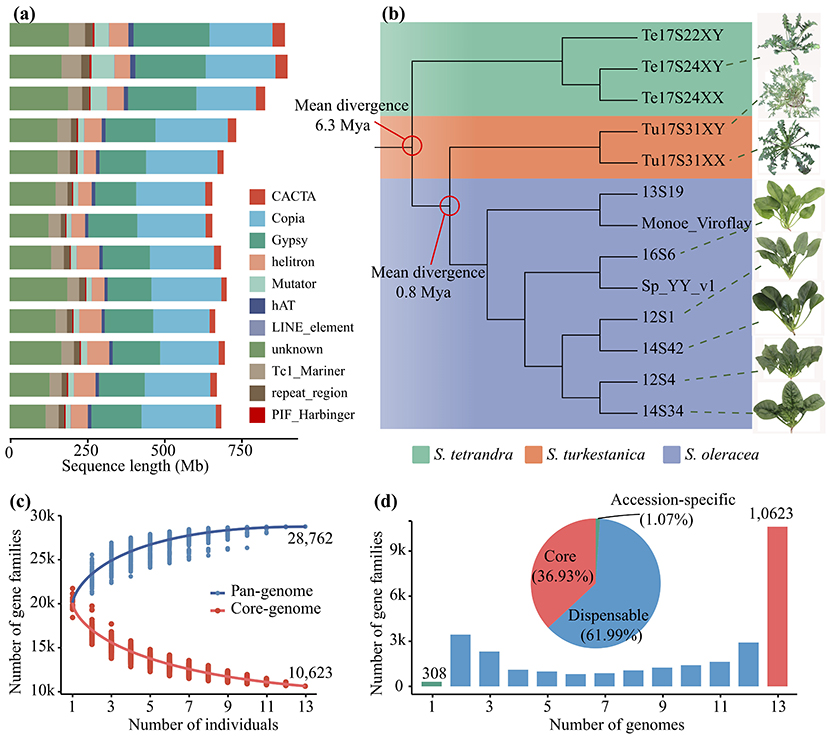
Figure 1 Phylogenetic relationships, genomic components and pan-genome of wild and cultivated spinach species
Comparing 13 spinach genome sequences, the study identified a total of 193,661 pan-structural variants (pan-SV), of which 43.65% were insertions and 37.24% were deletions. Population analysis revealed that pan-SV could classify the 455 Spinacia accesions into three subgroups, namely the wild species S. tetrandra, the wild species S. turkestanica, and the cultivated species, among which the cultivated species could be divided into the Asian type spinach and the European type spinach (Fig. 2), and the above results indicated that SV was closely related to species differentiation. In addition, pan-SV helps us to understand the direction of spinach evolution, for example, we found a 260 bp deletion that is closely linked to spinach sex, but it is actually an insertion on the X chromosome in spinach evolution (Fig. 2).
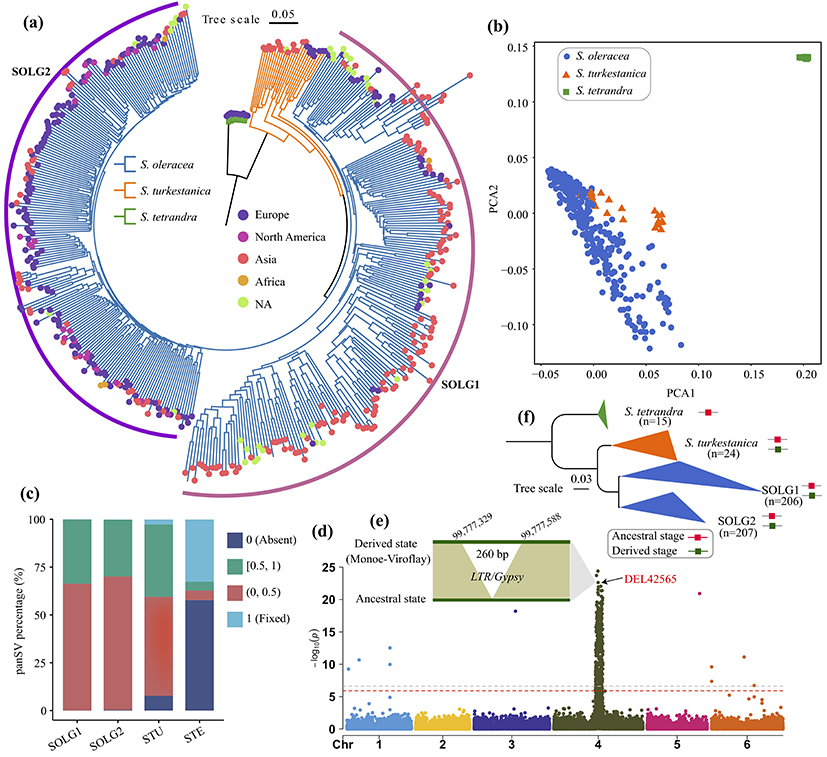
Figure 2 The utility of the pan-SV for population and GWAS analysis.
During sex chromosome evolution, the Y (W) chromosome tends to undergo degenerative phenomena, such as reduced gene expression, loss, or even resulting in a shorter Y chromosome (e.g., human Y chromosome). In this study, we found that in spinach sex chromosome evolution, SV was more inclined to occur on the Y chromosome, while the Y chromosome gene expression trend was lower than that of its alleles on the X chromosome. The above results suggest that SV may reduce Y chromosome gene expression and promote spinach Y chromosome degeneration (Fig. 3).
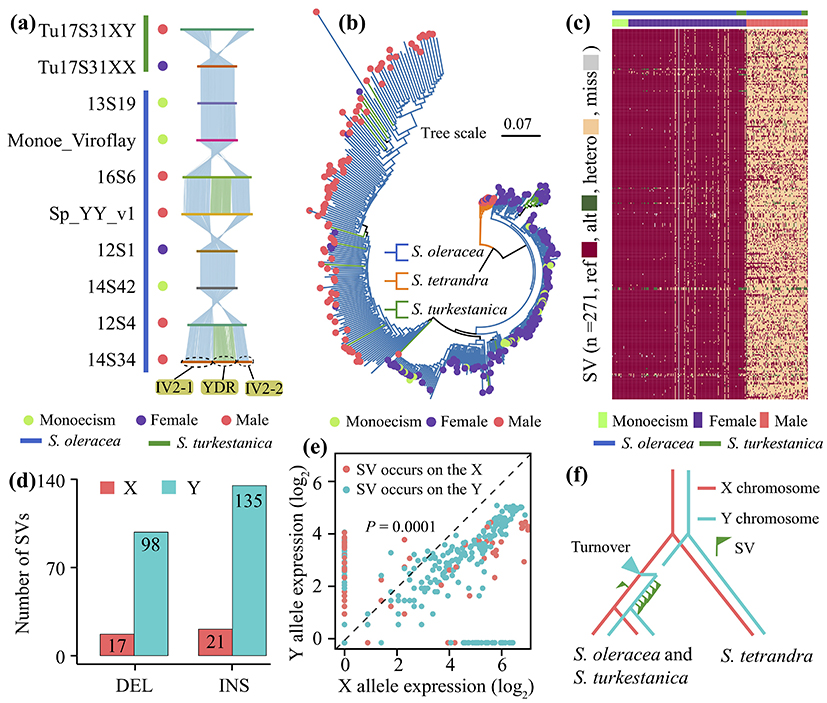
Figure 3 The evolution of sex-linked region (SLR) in spinach
In addition, this study also unearthed a large number of selected SVs that might be involved in the domestication of important agronomic traits, e.g., leaf margin type, flowering time, and seed dormancy. Meanwhile, it was further demonstrated that SVs could capture more genetic information compared with SNPs and had higher accuracy in genomic selection breeding. In conclusion, this study not only provides new insights into spinach sex chromosome evolution, but also lays an important foundation for mining spinach genes related to important agronomic traits and accelerating spinach molecular design breeding.
Assistant Researcher Hongbing She and Researcher Zhiyuan Liu from the IVF-CAAS, are the co-first authors of this paper; Researcher Wei Qian and Researcher Xiaowu Wang from the IVF-CAAS, are the co-corresponding authors. This work was supported by the Central Public-interest Scientific Institution Basal Research Fund (IVF-BRF2023002), Youth innovation Program of Chinese Academy of Agricultural Sciences (Y2023QC07), Chinese Academy of Agricultural Sciences Innovation Project (CAAS-ASTIP-IVFCAAS), and China Agricultural Research System (CARS-23-A-17).
More information can be found through the link: https://onlinelibrary.wiley.com/doi/full/10.1111/pbi.14433
By Hongbing She (shehongbing@caas.cn)