Recently, the Vegetable Molecular Design Breeding Team, in collaboration with the Potato Genetics Breeding and Cultivation Research Team at the Institute of Vegetables and Flowers, investigated the genetic characteristics of F1 segregating populations in outcross species or species with long generation times, and developed a specialised algorithm/tool called OcBSA to speed up gene discovery and advance molecular breeding in these species. The research, entitled "OcBSA: an NGS-based Bulk Segregant Analysis Tool for Outcross Populations", has been published in Molecular Plant.

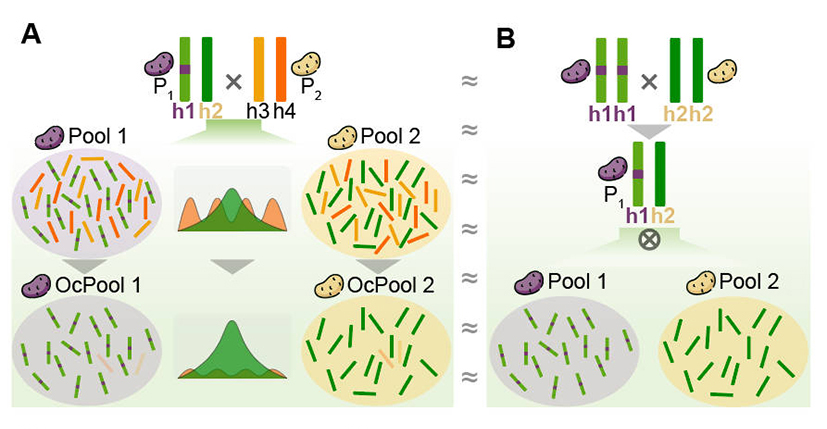
Many species, including vegetables, tubers, field crops, flowers, fruit trees and forest trees, are outcrossed. However, due to the high heterozygosity of their genomes, self-incompatibility or long generation times, the construction of inbred lines for genetic breeding research is challenging and hinders the genetic dissection of traits. Traits segregate in F1 individuals of these species. Effective use of F1 populations for rapid and accurate mapping of trait-controlling genes is a challenge for scientists in related fields of research. OcBSA simplifies the inheritance problem of four haplotypes from the two heterozygous parents of F1 populations by removing two informationless haplotypes, allowing efficient and accurate mapping of target trait control genes in F1 segregating populations. Extensive analysis of simulated and real data from eight species demonstrates the efficiency, accuracy and broad applicability of OcBSA. It is expected that OcBSA will significantly advance genetic analysis in outcrossing and long-generation species, thereby advancing molecular design and breeding research in these species.
This study was supported by the State Key Laboratory of Vegetable Biobreeding, the National Natural Science Foundation of China, the Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences, and the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture and Rural Affairs, P.R. China.
The link to the paper: https://doi.org/10.1016/j.molp.2024.02.011
By ChengFeng(chengfeng@caas.cn)