Recently, the vegetable pathogen research group of the IVF-CAAS, published a research paper titled "Deciphering the role of rhizosphere microbiota in modulating disease resistance in cabbage varieties" in the internationally famous journal Microbiome (IF=13.48, top 5% in the microbiology field). This study reveals for the first time that disease resistance genes (R genes) can reshape the rhizosphere microbiota of plants. By recruiting beneficial microorganisms as core microbial populations, a more stable and complex microbial network structure is constructed, thereby enhancing plant disease resistance through a new mechanism. This research provides new theoretical support for establishing new strategies for plant disease prevention and control.

In recent years, multiple studies have found that plants can regulate microbial communities through M genes (microbiome-shape) to enhance their disease resistance. Plant resistance genes (R genes) are the most critical genes for plants to resist diseases. However, there have been no reports on whether R genes can function as M genes to reshape microbial communities and subsequently inhibit the occurrence of diseases. Cabbage Fusarium wilt (CFW) is a serious soil-borne disease that threatens cabbage production, and the resistance of cabbage resistant varieties to CFW is determined by a single NBS-type resistance gene (FOC1). Previously, the cabbage research group of IVFCAAS introduced the FOC1 gene into Zhonggan 21 (ZG, highly susceptible to CFW), resulting in the development of a new variety, Zhonggan 21 (YR, highly resistant to CFW). ZG and YR have highly similar genetic backgrounds, and the difference in resistance to CFW is determined by just one R gene, making them excellent materials for studying the relationship between R genes and root microbial communities. This study first identified the differences in the composition of root microbial communities between ZG and YR through microbial community analysis. It was found that the root microbial species composition of the resistant variety was richer and formed a more complex and stable microbial network structure. Further research indicated that the R gene reshaped the rhizosphere microbial community and, by recruiting beneficial microorganisms, formed a highly stable network centered around beneficial microbes, thereby better resisting pathogen invasion. On this basis, by integrating data from microbiomes, cultivation groups, and genomics, key species (hub taxa) in the core microbial community of the resistant variety's rhizosphere were isolated and identified, including Pseudomonas brassicacearum NA13. In vitro plate inhibition and in vivo pot inoculation experiments demonstrated that this strain can suppress CFW by directly inhibiting it and inducing plant resistance. Analysis of the biocontrol mechanism of the biocontrol bacterium NA13 showed that NA13 can enhance plant resistance to pathogens by regulating the expression of key genes in plant hormone pathways, such as ethylene, jasmonic acid, and salicylic acid. Further experiments indicated that NA13 can also significantly improve crop resistance to other soil-borne diseases, such as root knot disease and bacterial wilt, showing great potential for biocontrol applications.
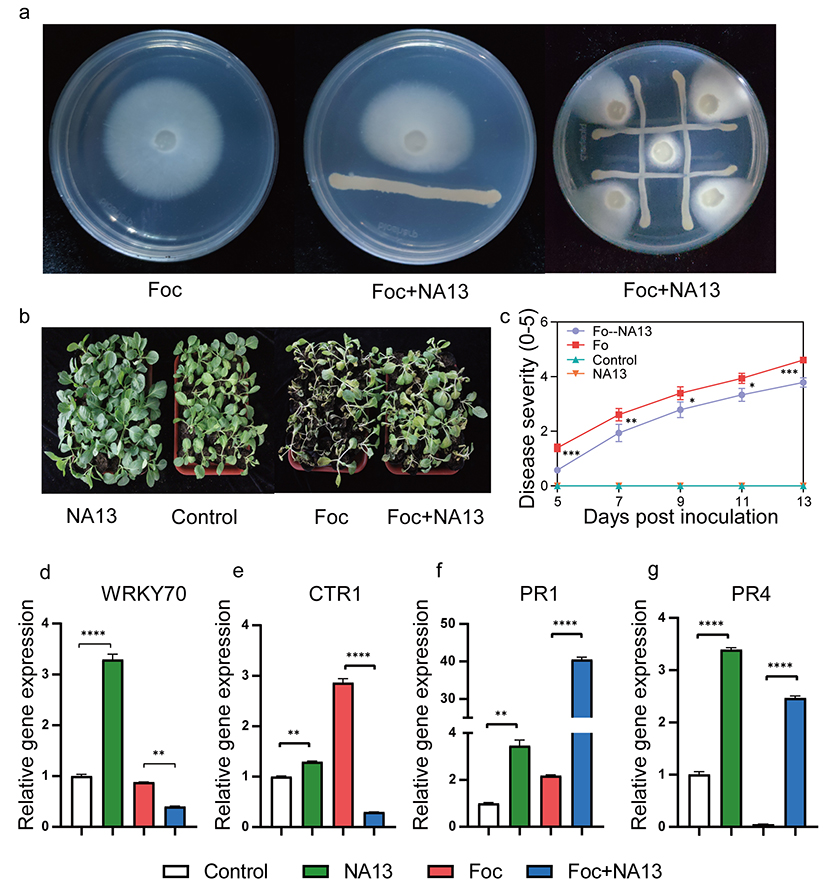
Figure 1: The inhibitory effect of the core microbial community NA13 on CFW
This study uncovered a new role of plant R genes beyond their traditional disease resistance function, demonstrating that they can enhance crop disease resistance by reshaping the rhizosphere microbial community. This work provides theoretical support for the organic integration of disease-resistant breeding and microbial community management, while also offering new insights for the exploration of combined resistant varieties and the development of synthetic microbiomes (Syncom) to create novel biocontrol agents.
Ping Xingxing, a graduate master's student from the Vegetable Institute (currently a PhD student at Nankai University), is the first author of the paper. His master's supervisor, Professor Ling Jian, along with Professor Xie Bingyan and Mao Zhenchuan, are the corresponding authors, with the IVFCAAS as the corresponding institution. Professor Lv Honghao from the cabbage research group at the IVFCAAS provided significant support for this study. This research was funded by the National Key Research and Development Program (2022YFD1401200), the National Key Laboratory of Vegetable Biotechnology, the National Natural Science Foundation (31571962), and other projects.
The link for the paper: https://doi.org/10.1186/s40168-024-01883-0